Streaming Columnar Data with Apache Arrow
Over the past couple weeks, Nong Li and I added a streaming binary format to Apache Arrow, accompanying the existing random access / IPC file format. We have implementations in Java and C++, plus Python bindings. In this post, I explain how the format works and show how you can achieve very high data throughput to pandas DataFrames.
Columnar streaming data
A common question I get about using Arrow is the high cost of transposing large tabular datasets from record- or row-oriented format to column-oriented format. For a multi-gigabyte dataset, transposing in memory or on disk may be prohibitive.
For streaming data, whether the source data is row-oriented or column-oriented memory layout, one option is to send small batches of rows, each internally having a columnar memory layout.
In Apache Arrow, an in-memory columnar array collection representing a chunk of a table is called a record batch. Multiple record batches can be collected to represent a single logical table data structure.
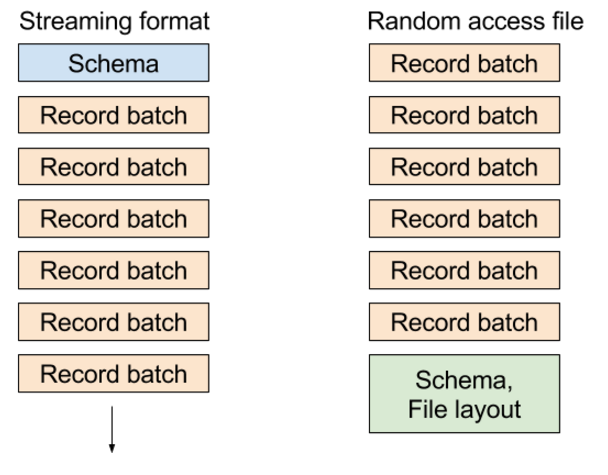
In the existing “random access” file format, we write metadata containing the table schema and block locations at the end of the file, enabling you to select any record batch or any column in the dataset very cheaply. In the streaming format, we send a series of messages: the schema followed by one or more record batches.
The different formats look roughly like this diagram:
Streaming data in PyArrow: Usage
To show you how this works, I generate an example dataset representing a single streaming chunk:
import time
import numpy as np
import pandas as pd
import pyarrow as pa
def generate_data(total_size, ncols):
nrows = int(total_size / ncols / np.dtype('float64').itemsize)
return pd.DataFrame({
'c' + str(i): np.random.randn(nrows)
for i in range(ncols)
})Now, suppose we want to write 1 gigabyte of data composed of chunks that are 1 megabyte each, so 1024 chunks. First, let’s create 1MB DataFrame with 16 columns:
KILOBYTE = 1 << 10
MEGABYTE = KILOBYTE * KILOBYTE
DATA_SIZE = 1024 * MEGABYTE
NCOLS = 16
df = generate_data(MEGABYTE, NCOLS)Then, I convert this to a pyarrow.RecordBatch:
batch = pa.RecordBatch.from_pandas(df)Now, I create an output stream that writes to RAM and create a StreamWriter:
sink = pa.InMemoryOutputStream()
stream_writer = pa.StreamWriter(sink, batch.schema)Then, we write the 1024 chunks composing the 1 GB dataset:
for i in range(DATA_SIZE // MEGABYTE):
stream_writer.write_batch(batch)Since we wrote to RAM, we can get the entire stream as a single buffer:
In [13]: source = sink.get_result()
In [14]: source
Out[14]: <pyarrow.io.Buffer at 0x7f2df7118f80>
In [15]: source.size
Out[15]: 1074750744Since this data is in memory, reading back Arrow record batches is a zero-copy operation. I open a StreamReader, read back the data as a pyarrow.Table, and then convert to a pandas DataFrame:
In [16]: reader = pa.StreamReader(source)
In [17]: table = reader.read_all()
In [18]: table
Out[18]: <pyarrow.table.Table at 0x7fae8281f6f0>
In [19]: df = table.to_pandas()
In [20]: df.memory_usage().sum()
Out[20]: 1073741904This is all very nice, but you may have some questions. How fast is it? How does the stream chunk size affect the absolute performance to obtain the pandas DataFrame?
Streaming data performance
As the streaming chunksize grows smaller, the cost to reconstruct a contiguous columnar pandas DataFrame increases because of cache-inefficient memory access patterns. There is also some overhead from manipulating the C++ container data structures around the arrays and their memory buffers.
With a 1 MB as above, on my laptop (Quad-core Xeon E3-1505M) I have:
In [20]: %timeit pa.StreamReader(source).read_all().to_pandas()
10 loops, best of 3: 129 ms per loopThis is an effective throughput of 7.75 GB/s to reconstruct a 1GB DataFrame from 1024 1MB chunks. What happens when we use larger and smaller chunks? Here are the results
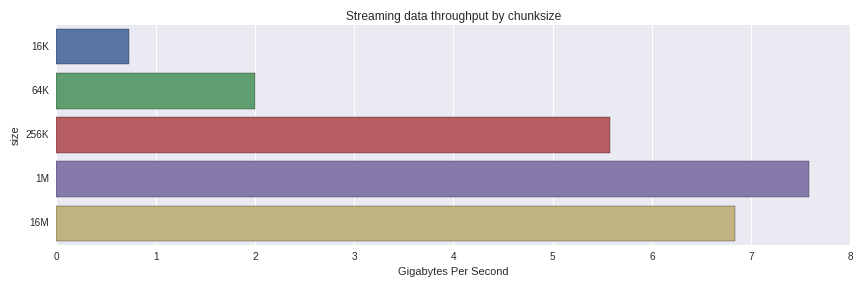
The performance degrades significantly from 256K to 64K chunks. I was surprised to see that 1MB chunks were faster than 16MB ones; it would be worth a more thorough investigation to understand whether that is normal variance or something else going on.
In the current iteration of the format, the data is not being compressed at all, so the in-memory and on-the-wire size are about the same. Compression may be added to the format as an option in the future.
Summary
Streaming columnar data can be an efficient way to transmit large datasets to columnar analytics tools like pandas using small chunks. Data services using row-oriented storage can transpose and stream small data chunks that are more friendly to your CPU’s L2 and L3 caches.
Full benchmarking code
import time
import numpy as np
import pandas as pd
import pyarrow as pa
def generate_data(total_size, ncols):
nrows = total_size / ncols / np.dtype('float64').itemsize
return pd.DataFrame({
'c' + str(i): np.random.randn(nrows)
for i in range(ncols)
})
KILOBYTE = 1 << 10
MEGABYTE = KILOBYTE * KILOBYTE
DATA_SIZE = 1024 * MEGABYTE
NCOLS = 16
def get_timing(f, niter):
start = time.clock_gettime(time.CLOCK_REALTIME)
for i in range(niter):
f()
return (time.clock_gettime(time.CLOCK_REALTIME) - start) / NITER
def read_as_dataframe(klass, source):
reader = klass(source)
table = reader.read_all()
return table.to_pandas()
NITER = 5
results = []
CHUNKSIZES = [16 * KILOBYTE, 64 * KILOBYTE, 256 * KILOBYTE, MEGABYTE, 16 * MEGABYTE]
for chunksize in CHUNKSIZES:
nchunks = DATA_SIZE // chunksize
batch = pa.RecordBatch.from_pandas(generate_data(chunksize, NCOLS))
sink = pa.InMemoryOutputStream()
stream_writer = pa.StreamWriter(sink, batch.schema)
for i in range(nchunks):
stream_writer.write_batch(batch)
source = sink.get_result()
elapsed = get_timing(lambda: read_as_dataframe(pa.StreamReader, source), NITER)
result = (chunksize, elapsed)
print(result)
results.append(result)