Extreme IO performance with parallel Apache Parquet in Python
In this post, I show how Parquet can encode very large datasets in a small file footprint, and how we can achieve data throughput significantly exceeding disk IO bandwidth by exploiting parallelism (multithreading).
Apache Parquet: Top performer on low-entropy data
As you can read in the Apache Parquet format specification, the format features multiple layers of encoding to achieve small file size, among them:
- Dictionary encoding (similar to how
pandas.Categoricalrepresents data, but they aren’t equivalent concepts) - Data page compression (Snappy, Gzip, LZO, or Brotli)
- Run-length encoding (for null indicators and dictionary indices) and integer bit-packing
To give you an idea of how this works, let’s consider the dataset:
['banana', 'banana', 'banana', 'banana', 'banana', 'banana',
'banana', 'banana', 'apple', 'apple', 'apple']Almost all Parquet implementations dictionary encode by default. So the first pass encoding becomes:
dictionary: ['banana', 'apple']
indices: [0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1]The dictionary indices are further run-length encoded:
dictionary: ['banana', 'apple']
indices (RLE): [(8, 0), (3, 1)]Working backwards, you can easily reconstruct the original dense array of strings.
In my prior blog post, I created a dataset that compresses very well with this style of encoding. When writing with pyarrow, we can turn on and off dictionary encoding (which is on by default) to see how it impacts file size:
import pyarrow.parquet as pq
pq.write_table(dataset, out_path, use_dictionary=True,
compression='snappy)With a dataset that occupies 1 gigabyte (1024 MB) in a pandas.DataFrame, with Snappy compression and dictionary encoding, it occupies an amazing 1.436 MB, small enough to fit on an old-school floppy disk. Without dictionary encoding, it occupies 44.4 MB.
Parallel reads in parquet-cpp via PyArrow
In parquet-cpp, the C++ implementation of Apache Parquet, which we’ve made available to Python in PyArrow, we recently added parallel column reads.
To try this out, install PyArrow from conda-forge:
conda install pyarrow -c conda-forgeNow, when reading a Parquet file, use the nthreads argument:
import pyarrow.parquet as pq
table = pq.read_table(file_path, nthreads=4)For low entropy data, decompression and decoding becomes CPU-bound. Because we are doing all the work in C++, we are not burdened by the concurrency issues of the GIL and thus can achieve a significant speed boost. See the results I achieved reading a 1 GB dataset to a pandas DataFrame on my quad-core laptop (Xeon E3-1505M, NVMe SSD):
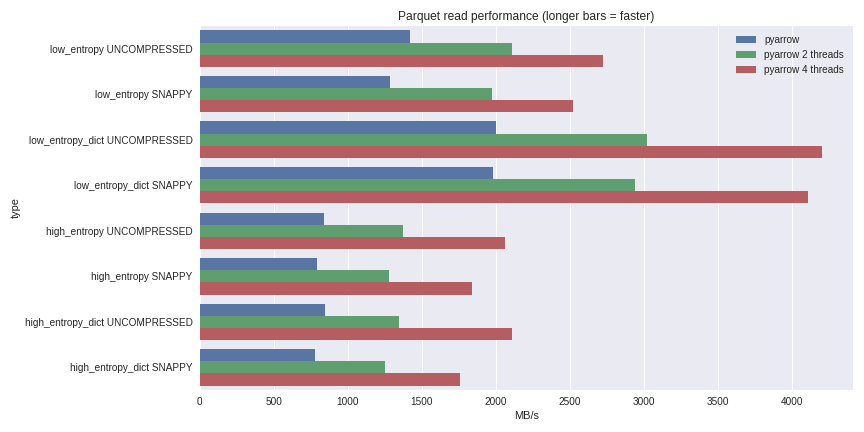
Click here for full benchmarking script
I included performance both for the dictionary-encoded and non-dictionary encoded cases. For low entropy data, even though the files are all small (~1.5 MB with dictionaries and ~45 MB without), the impact of dictionary encoding on performance is substantial. With 4 threads, the performance reading into pandas breaks through an amazing 4 GB/s. This is much faster than Feather format or other alternatives I’ve seen.
Conclusions
With the 1.0 release of parquet-cpp (Apache Parquet in C++) on the horizon, it’s great to see this kind of IO performance made available to the Python user base.
Since all of the underlying machinery here is implemented in C++, other languages (such as R) can build interfaces to Apache Arrow (the common columnar data structures) and parquet-cpp. The Python bindings are a lightweight wrapper on top of the underlying libarrow and libparquet C++ libraries.