10 Data Aggregation and Group Operations
Categorizing a dataset and applying a function to each group, whether an aggregation or transformation, can be a critical component of a data analysis workflow. After loading, merging, and preparing a dataset, you may need to compute group statistics or possibly pivot tables for reporting or visualization purposes. pandas provides a versatile groupby interface, enabling you to slice, dice, and summarize datasets in a natural way.
One reason for the popularity of relational databases and SQL (which stands for “structured query language”) is the ease with which data can be joined, filtered, transformed, and aggregated. However, query languages like SQL impose certain limitations on the kinds of group operations that can be performed. As you will see, with the expressiveness of Python and pandas, we can perform quite complex group operations by expressing them as custom Python functions that manipulate the data associated with each group. In this chapter, you will learn how to:
Split a pandas object into pieces using one or more keys (in the form of functions, arrays, or DataFrame column names)
Calculate group summary statistics, like count, mean, or standard deviation, or a user-defined function
Apply within-group transformations or other manipulations, like normalization, linear regression, rank, or subset selection
Compute pivot tables and cross-tabulations
Perform quantile analysis and other statistical group analyses
Time-based aggregation of time series data, a special use case of groupby, is referred to as resampling in this book and will receive separate treatment in Ch 11: Time Series.
As with the rest of the chapters, we start by importing NumPy and pandas:
In [12]: import numpy as np
In [13]: import pandas as pd10.1 How to Think About Group Operations
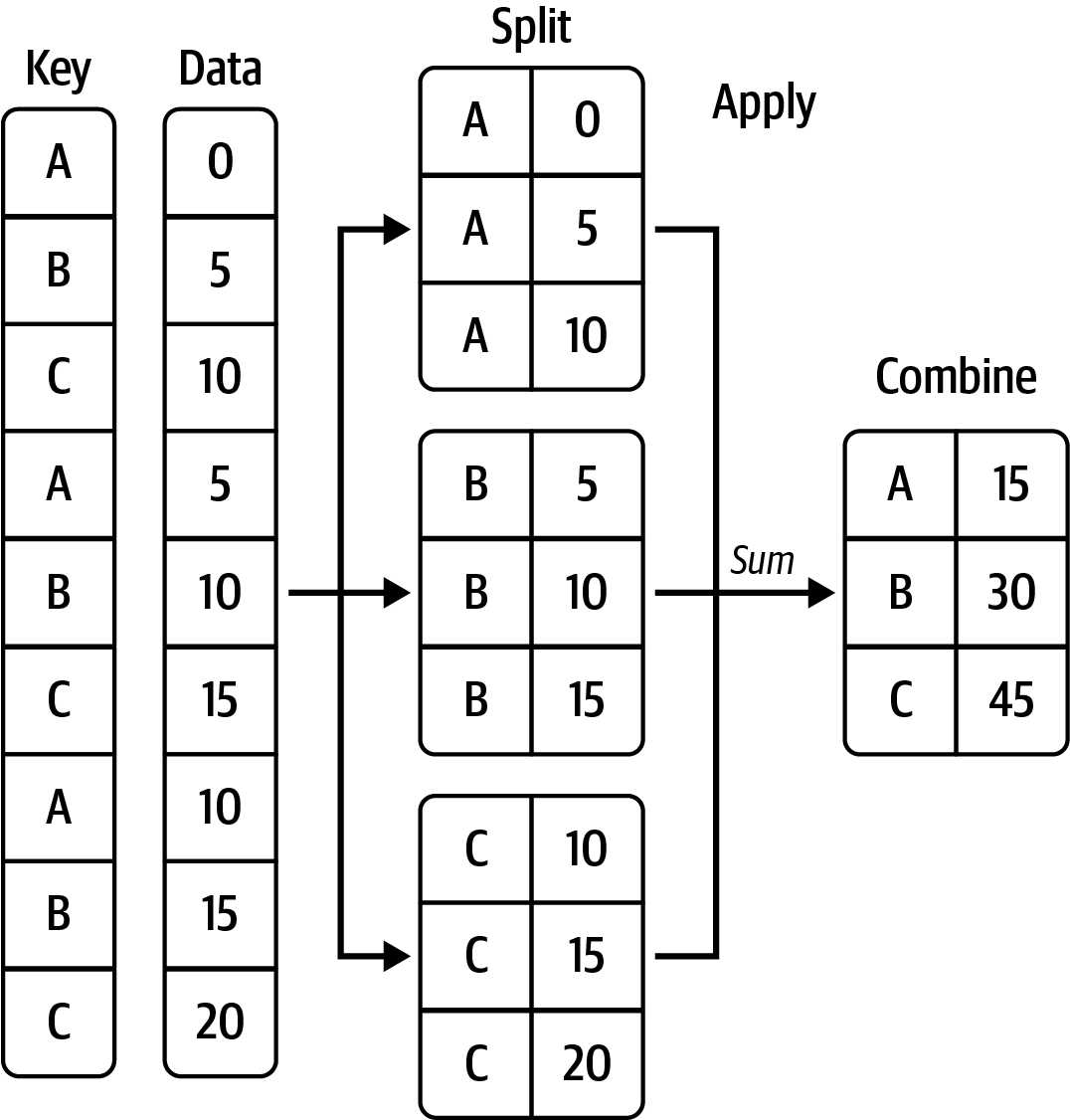
Hadley Wickham, an author of many popular packages for the R programming language, coined the term split-apply-combine for describing group operations. In the first stage of the process, data contained in a pandas object, whether a Series, DataFrame, or otherwise, is split into groups based on one or more keys that you provide. The splitting is performed on a particular axis of an object. For example, a DataFrame can be grouped on its rows (axis="index") or its columns (axis="columns"). Once this is done, a function is applied to each group, producing a new value. Finally, the results of all those function applications are combined into a result object. The form of the resulting object will usually depend on what’s being done to the data. See Figure 10.1 for a mockup of a simple group aggregation.
Each grouping key can take many forms, and the keys do not have to be all of the same type:
A list or array of values that is the same length as the axis being grouped
A value indicating a column name in a DataFrame
A dictionary or Series giving a correspondence between the values on the axis being grouped and the group names
A function to be invoked on the axis index or the individual labels in the index
Note that the latter three methods are shortcuts for producing an array of values to be used to split up the object. Don’t worry if this all seems abstract. Throughout this chapter, I will give many examples of all these methods. To get started, here is a small tabular dataset as a DataFrame:
In [14]: df = pd.DataFrame({"key1" : ["a", "a", None, "b", "b", "a", None],
....: "key2" : pd.Series([1, 2, 1, 2, 1, None, 1],
....: dtype="Int64"),
....: "data1" : np.random.standard_normal(7),
....: "data2" : np.random.standard_normal(7)})
In [15]: df
Out[15]:
key1 key2 data1 data2
0 a 1 -0.204708 0.281746
1 a 2 0.478943 0.769023
2 None 1 -0.519439 1.246435
3 b 2 -0.555730 1.007189
4 b 1 1.965781 -1.296221
5 a <NA> 1.393406 0.274992
6 None 1 0.092908 0.228913Suppose you wanted to compute the mean of the data1 column using the labels from key1. There are a number of ways to do this. One is to access data1 and call groupby with the column (a Series) at key1:
In [16]: grouped = df["data1"].groupby(df["key1"])
In [17]: grouped
Out[17]: <pandas.core.groupby.generic.SeriesGroupBy object at 0x17b7913f0>This grouped variable is now a special "GroupBy" object. It has not actually computed anything yet except for some intermediate data about the group key df["key1"]. The idea is that this object has all of the information needed to then apply some operation to each of the groups. For example, to compute group means we can call the GroupBy’s mean method:
In [18]: grouped.mean()
Out[18]:
key1
a 0.555881
b 0.705025
Name: data1, dtype: float64Later in Data Aggregation, I'll explain more about what happens when you call .mean(). The important thing here is that the data (a Series) has been aggregated by splitting the data on the group key, producing a new Series that is now indexed by the unique values in the key1 column. The result index has the name "key1" because the DataFrame column df["key1"] did.
If instead we had passed multiple arrays as a list, we'd get something different:
In [19]: means = df["data1"].groupby([df["key1"], df["key2"]]).mean()
In [20]: means
Out[20]:
key1 key2
a 1 -0.204708
2 0.478943
b 1 1.965781
2 -0.555730
Name: data1, dtype: float64Here we grouped the data using two keys, and the resulting Series now has a hierarchical index consisting of the unique pairs of keys observed:
In [21]: means.unstack()
Out[21]:
key2 1 2
key1
a -0.204708 0.478943
b 1.965781 -0.555730In this example, the group keys are all Series, though they could be any arrays of the right length:
In [22]: states = np.array(["OH", "CA", "CA", "OH", "OH", "CA", "OH"])
In [23]: years = [2005, 2005, 2006, 2005, 2006, 2005, 2006]
In [24]: df["data1"].groupby([states, years]).mean()
Out[24]:
CA 2005 0.936175
2006 -0.519439
OH 2005 -0.380219
2006 1.029344
Name: data1, dtype: float64Frequently, the grouping information is found in the same DataFrame as the data you want to work on. In that case, you can pass column names (whether those are strings, numbers, or other Python objects) as the group keys:
In [25]: df.groupby("key1").mean()
Out[25]:
key2 data1 data2
key1
a 1.5 0.555881 0.441920
b 1.5 0.705025 -0.144516
In [26]: df.groupby("key2").mean(numeric_only=True)
Out[26]:
data1 data2
key2
1 0.333636 0.115218
2 -0.038393 0.888106
In [27]: df.groupby(["key1", "key2"]).mean()
Out[27]:
data1 data2
key1 key2
a 1 -0.204708 0.281746
2 0.478943 0.769023
b 1 1.965781 -1.296221
2 -0.555730 1.007189You may notice that in the second case, it is necessary to pass numeric_only=True because the key1 column is not numeric and thus cannot be aggregated with mean().
Regardless of the objective in using groupby, a generally useful GroupBy method is size, which returns a Series containing group sizes:
In [28]: df.groupby(["key1", "key2"]).size()
Out[28]:
key1 key2
a 1 1
2 1
b 1 1
2 1
dtype: int64Note that any missing values in a group key are excluded from the result by default. This behavior can be disabled by passing dropna=False to groupby:
In [29]: df.groupby("key1", dropna=False).size()
Out[29]:
key1
a 3
b 2
NaN 2
dtype: int64
In [30]: df.groupby(["key1", "key2"], dropna=False).size()
Out[30]:
key1 key2
a 1 1
2 1
<NA> 1
b 1 1
2 1
NaN 1 2
dtype: int64A group function similar in spirit to size is count, which computes the number of nonnull values in each group:
In [31]: df.groupby("key1").count()
Out[31]:
key2 data1 data2
key1
a 2 3 3
b 2 2 2Iterating over Groups
The object returned by groupby supports iteration, generating a sequence of 2-tuples containing the group name along with the chunk of data. Consider the following:
In [32]: for name, group in df.groupby("key1"):
....: print(name)
....: print(group)
....:
a
key1 key2 data1 data2
0 a 1 -0.204708 0.281746
1 a 2 0.478943 0.769023
5 a <NA> 1.393406 0.274992
b
key1 key2 data1 data2
3 b 2 -0.555730 1.007189
4 b 1 1.965781 -1.296221In the case of multiple keys, the first element in the tuple will be a tuple of key values:
In [33]: for (k1, k2), group in df.groupby(["key1", "key2"]):
....: print((k1, k2))
....: print(group)
....:
('a', 1)
key1 key2 data1 data2
0 a 1 -0.204708 0.281746
('a', 2)
key1 key2 data1 data2
1 a 2 0.478943 0.769023
('b', 1)
key1 key2 data1 data2
4 b 1 1.965781 -1.296221
('b', 2)
key1 key2 data1 data2
3 b 2 -0.55573 1.007189Of course, you can choose to do whatever you want with the pieces of data. A recipe you may find useful is computing a dictionary of the data pieces as a one-liner:
In [34]: pieces = {name: group for name, group in df.groupby("key1")}
In [35]: pieces["b"]
Out[35]:
key1 key2 data1 data2
3 b 2 -0.555730 1.007189
4 b 1 1.965781 -1.296221By default groupby groups on axis="index", but you can group on any of the other axes. For example, we could group the columns of our example df here by whether they start with "key" or "data":
In [36]: grouped = df.groupby({"key1": "key", "key2": "key",
....: "data1": "data", "data2": "data"}, axis="columns")We can print out the groups like so:
In [37]: for group_key, group_values in grouped:
....: print(group_key)
....: print(group_values)
....:
data
data1 data2
0 -0.204708 0.281746
1 0.478943 0.769023
2 -0.519439 1.246435
3 -0.555730 1.007189
4 1.965781 -1.296221
5 1.393406 0.274992
6 0.092908 0.228913
key
key1 key2
0 a 1
1 a 2
2 None 1
3 b 2
4 b 1
5 a <NA>
6 None 1Selecting a Column or Subset of Columns
Indexing a GroupBy object created from a DataFrame with a column name or array of column names has the effect of column subsetting for aggregation. This means that:
df.groupby("key1")["data1"]
df.groupby("key1")[["data2"]]are conveniences for:
df["data1"].groupby(df["key1"])
df[["data2"]].groupby(df["key1"])Especially for large datasets, it may be desirable to aggregate only a few columns. For example, in the preceding dataset, to compute the means for just the data2 column and get the result as a DataFrame, we could write:
In [38]: df.groupby(["key1", "key2"])[["data2"]].mean()
Out[38]:
data2
key1 key2
a 1 0.281746
2 0.769023
b 1 -1.296221
2 1.007189The object returned by this indexing operation is a grouped DataFrame if a list or array is passed, or a grouped Series if only a single column name is passed as a scalar:
In [39]: s_grouped = df.groupby(["key1", "key2"])["data2"]
In [40]: s_grouped
Out[40]: <pandas.core.groupby.generic.SeriesGroupBy object at 0x17b8356c0>
In [41]: s_grouped.mean()
Out[41]:
key1 key2
a 1 0.281746
2 0.769023
b 1 -1.296221
2 1.007189
Name: data2, dtype: float64Grouping with Dictionaries and Series
Grouping information may exist in a form other than an array. Let’s consider another example DataFrame:
In [42]: people = pd.DataFrame(np.random.standard_normal((5, 5)),
....: columns=["a", "b", "c", "d", "e"],
....: index=["Joe", "Steve", "Wanda", "Jill", "Trey"])
In [43]: people.iloc[2:3, [1, 2]] = np.nan # Add a few NA values
In [44]: people
Out[44]:
a b c d e
Joe 1.352917 0.886429 -2.001637 -0.371843 1.669025
Steve -0.438570 -0.539741 0.476985 3.248944 -1.021228
Wanda -0.577087 NaN NaN 0.523772 0.000940
Jill 1.343810 -0.713544 -0.831154 -2.370232 -1.860761
Trey -0.860757 0.560145 -1.265934 0.119827 -1.063512Now, suppose I have a group correspondence for the columns and want to sum the columns by group:
In [45]: mapping = {"a": "red", "b": "red", "c": "blue",
....: "d": "blue", "e": "red", "f" : "orange"}Now, you could construct an array from this dictionary to pass to groupby, but instead we can just pass the dictionary (I included the key "f" to highlight that unused grouping keys are OK):
In [46]: by_column = people.groupby(mapping, axis="columns")
In [47]: by_column.sum()
Out[47]:
blue red
Joe -2.373480 3.908371
Steve 3.725929 -1.999539
Wanda 0.523772 -0.576147
Jill -3.201385 -1.230495
Trey -1.146107 -1.364125The same functionality holds for Series, which can be viewed as a fixed-size mapping:
In [48]: map_series = pd.Series(mapping)
In [49]: map_series
Out[49]:
a red
b red
c blue
d blue
e red
f orange
dtype: object
In [50]: people.groupby(map_series, axis="columns").count()
Out[50]:
blue red
Joe 2 3
Steve 2 3
Wanda 1 2
Jill 2 3
Trey 2 3Grouping with Functions
Using Python functions is a more generic way of defining a group mapping compared with a dictionary or Series. Any function passed as a group key will be called once per index value (or once per column value if using axis="columns"), with the return values being used as the group names. More concretely, consider the example DataFrame from the previous section, which has people’s first names as index values. Suppose you wanted to group by name length. While you could compute an array of string lengths, it's simpler to just pass the len function:
In [51]: people.groupby(len).sum()
Out[51]:
a b c d e
3 1.352917 0.886429 -2.001637 -0.371843 1.669025
4 0.483052 -0.153399 -2.097088 -2.250405 -2.924273
5 -1.015657 -0.539741 0.476985 3.772716 -1.020287Mixing functions with arrays, dictionaries, or Series is not a problem, as everything gets converted to arrays internally:
In [52]: key_list = ["one", "one", "one", "two", "two"]
In [53]: people.groupby([len, key_list]).min()
Out[53]:
a b c d e
3 one 1.352917 0.886429 -2.001637 -0.371843 1.669025
4 two -0.860757 -0.713544 -1.265934 -2.370232 -1.860761
5 one -0.577087 -0.539741 0.476985 0.523772 -1.021228Grouping by Index Levels
A final convenience for hierarchically indexed datasets is the ability to aggregate using one of the levels of an axis index. Let's look at an example:
In [54]: columns = pd.MultiIndex.from_arrays([["US", "US", "US", "JP", "JP"],
....: [1, 3, 5, 1, 3]],
....: names=["cty", "tenor"])
In [55]: hier_df = pd.DataFrame(np.random.standard_normal((4, 5)), columns=column
s)
In [56]: hier_df
Out[56]:
cty US JP
tenor 1 3 5 1 3
0 0.332883 -2.359419 -0.199543 -1.541996 -0.970736
1 -1.307030 0.286350 0.377984 -0.753887 0.331286
2 1.349742 0.069877 0.246674 -0.011862 1.004812
3 1.327195 -0.919262 -1.549106 0.022185 0.758363To group by level, pass the level number or name using the level keyword:
In [57]: hier_df.groupby(level="cty", axis="columns").count()
Out[57]:
cty JP US
0 2 3
1 2 3
2 2 3
3 2 310.2 Data Aggregation
Aggregations refer to any data transformation that produces scalar values from arrays. The preceding examples have used several of them, including mean, count, min, and sum. You may wonder what is going on when you invoke mean() on a GroupBy object. Many common aggregations, such as those found in Table 10.1, have optimized implementations. However, you are not limited to only this set of methods.
| Function name | Description |
|---|---|
any, all |
Return True if any (one or more values) or all non-NA values are "truthy" |
count |
Number of non-NA values |
cummin, cummax |
Cumulative minimum and maximum of non-NA values |
cumsum |
Cumulative sum of non-NA values |
cumprod |
Cumulative product of non-NA values |
first, last |
First and last non-NA values |
mean |
Mean of non-NA values |
median |
Arithmetic median of non-NA values |
min, max |
Minimum and maximum of non-NA values |
nth |
Retrieve value that would appear at position n with the data in sorted order |
ohlc |
Compute four "open-high-low-close" statistics for time series-like data |
prod |
Product of non-NA values |
quantile |
Compute sample quantile |
rank |
Ordinal ranks of non-NA values, like calling Series.rank |
size |
Compute group sizes, returning result as a Series |
sum |
Sum of non-NA values |
std, var |
Sample standard deviation and variance |
You can use aggregations of your own devising and additionally call any method that is also defined on the object being grouped. For example, the nsmallest Series method selects the smallest requested number of values from the data. While nsmallest is not explicitly implemented for GroupBy, we can still use it with a nonoptimized implementation. Internally, GroupBy slices up the Series, calls piece.nsmallest(n) for each piece, and then assembles those results into the result object:
In [58]: df
Out[58]:
key1 key2 data1 data2
0 a 1 -0.204708 0.281746
1 a 2 0.478943 0.769023
2 None 1 -0.519439 1.246435
3 b 2 -0.555730 1.007189
4 b 1 1.965781 -1.296221
5 a <NA> 1.393406 0.274992
6 None 1 0.092908 0.228913
In [59]: grouped = df.groupby("key1")
In [60]: grouped["data1"].nsmallest(2)
Out[60]:
key1
a 0 -0.204708
1 0.478943
b 3 -0.555730
4 1.965781
Name: data1, dtype: float64To use your own aggregation functions, pass any function that aggregates an array to the aggregate method or its short alias agg:
In [61]: def peak_to_peak(arr):
....: return arr.max() - arr.min()
In [62]: grouped.agg(peak_to_peak)
Out[62]:
key2 data1 data2
key1
a 1 1.598113 0.494031
b 1 2.521511 2.303410You may notice that some methods, like describe, also work, even though they are not aggregations, strictly speaking:
In [63]: grouped.describe()
Out[63]:
key2 data1 ...
count mean std min 25% 50% 75% max count mean ...
key1 ...
a 2.0 1.5 0.707107 1.0 1.25 1.5 1.75 2.0 3.0 0.555881 ... \
b 2.0 1.5 0.707107 1.0 1.25 1.5 1.75 2.0 2.0 0.705025 ...
data2
75% max count mean std min 25%
key1
a 0.936175 1.393406 3.0 0.441920 0.283299 0.274992 0.278369 \
b 1.335403 1.965781 2.0 -0.144516 1.628757 -1.296221 -0.720368
50% 75% max
key1
a 0.281746 0.525384 0.769023
b -0.144516 0.431337 1.007189
[2 rows x 24 columns]I will explain in more detail what has happened here in Apply: General split-apply-combine.
Custom aggregation functions are generally much slower than the optimized functions found in Table 10.1. This is because there is some extra overhead (function calls, data rearrangement) in constructing the intermediate group data chunks.
Column-Wise and Multiple Function Application
Let's return to the tipping dataset used in the last chapter. After loading it with pandas.read_csv, we add a tipping percentage column:
In [64]: tips = pd.read_csv("examples/tips.csv")
In [65]: tips.head()
Out[65]:
total_bill tip smoker day time size
0 16.99 1.01 No Sun Dinner 2
1 10.34 1.66 No Sun Dinner 3
2 21.01 3.50 No Sun Dinner 3
3 23.68 3.31 No Sun Dinner 2
4 24.59 3.61 No Sun Dinner 4Now I will add a tip_pct column with the tip percentage of the total bill:
In [66]: tips["tip_pct"] = tips["tip"] / tips["total_bill"]
In [67]: tips.head()
Out[67]:
total_bill tip smoker day time size tip_pct
0 16.99 1.01 No Sun Dinner 2 0.059447
1 10.34 1.66 No Sun Dinner 3 0.160542
2 21.01 3.50 No Sun Dinner 3 0.166587
3 23.68 3.31 No Sun Dinner 2 0.139780
4 24.59 3.61 No Sun Dinner 4 0.146808As you’ve already seen, aggregating a Series or all of the columns of a DataFrame is a matter of using aggregate (or agg) with the desired function or calling a method like mean or std. However, you may want to aggregate using a different function, depending on the column, or multiple functions at once. Fortunately, this is possible to do, which I’ll illustrate through a number of examples. First, I’ll group the tips by day and smoker:
In [68]: grouped = tips.groupby(["day", "smoker"])Note that for descriptive statistics like those in Table 10.1, you can pass the name of the function as a string:
In [69]: grouped_pct = grouped["tip_pct"]
In [70]: grouped_pct.agg("mean")
Out[70]:
day smoker
Fri No 0.151650
Yes 0.174783
Sat No 0.158048
Yes 0.147906
Sun No 0.160113
Yes 0.187250
Thur No 0.160298
Yes 0.163863
Name: tip_pct, dtype: float64If you pass a list of functions or function names instead, you get back a DataFrame with column names taken from the functions:
In [71]: grouped_pct.agg(["mean", "std", peak_to_peak])
Out[71]:
mean std peak_to_peak
day smoker
Fri No 0.151650 0.028123 0.067349
Yes 0.174783 0.051293 0.159925
Sat No 0.158048 0.039767 0.235193
Yes 0.147906 0.061375 0.290095
Sun No 0.160113 0.042347 0.193226
Yes 0.187250 0.154134 0.644685
Thur No 0.160298 0.038774 0.193350
Yes 0.163863 0.039389 0.151240Here we passed a list of aggregation functions to agg to evaluate independently on the data groups.
You don’t need to accept the names that GroupBy gives to the columns; notably, lambda functions have the name "<lambda>", which makes them hard to identify (you can see for yourself by looking at a function’s __name__ attribute). Thus, if you pass a list of (name, function) tuples, the first element of each tuple will be used as the DataFrame column names (you can think of a list of 2-tuples as an ordered mapping):
In [72]: grouped_pct.agg([("average", "mean"), ("stdev", np.std)])
Out[72]:
average stdev
day smoker
Fri No 0.151650 0.028123
Yes 0.174783 0.051293
Sat No 0.158048 0.039767
Yes 0.147906 0.061375
Sun No 0.160113 0.042347
Yes 0.187250 0.154134
Thur No 0.160298 0.038774
Yes 0.163863 0.039389With a DataFrame you have more options, as you can specify a list of functions to apply to all of the columns or different functions per column. To start, suppose we wanted to compute the same three statistics for the tip_pct and total_bill columns:
In [73]: functions = ["count", "mean", "max"]
In [74]: result = grouped[["tip_pct", "total_bill"]].agg(functions)
In [75]: result
Out[75]:
tip_pct total_bill
count mean max count mean max
day smoker
Fri No 4 0.151650 0.187735 4 18.420000 22.75
Yes 15 0.174783 0.263480 15 16.813333 40.17
Sat No 45 0.158048 0.291990 45 19.661778 48.33
Yes 42 0.147906 0.325733 42 21.276667 50.81
Sun No 57 0.160113 0.252672 57 20.506667 48.17
Yes 19 0.187250 0.710345 19 24.120000 45.35
Thur No 45 0.160298 0.266312 45 17.113111 41.19
Yes 17 0.163863 0.241255 17 19.190588 43.11As you can see, the resulting DataFrame has hierarchical columns, the same as you would get aggregating each column separately and using concat to glue the results together using the column names as the keys argument:
In [76]: result["tip_pct"]
Out[76]:
count mean max
day smoker
Fri No 4 0.151650 0.187735
Yes 15 0.174783 0.263480
Sat No 45 0.158048 0.291990
Yes 42 0.147906 0.325733
Sun No 57 0.160113 0.252672
Yes 19 0.187250 0.710345
Thur No 45 0.160298 0.266312
Yes 17 0.163863 0.241255As before, a list of tuples with custom names can be passed:
In [77]: ftuples = [("Average", "mean"), ("Variance", np.var)]
In [78]: grouped[["tip_pct", "total_bill"]].agg(ftuples)
Out[78]:
tip_pct total_bill
Average Variance Average Variance
day smoker
Fri No 0.151650 0.000791 18.420000 25.596333
Yes 0.174783 0.002631 16.813333 82.562438
Sat No 0.158048 0.001581 19.661778 79.908965
Yes 0.147906 0.003767 21.276667 101.387535
Sun No 0.160113 0.001793 20.506667 66.099980
Yes 0.187250 0.023757 24.120000 109.046044
Thur No 0.160298 0.001503 17.113111 59.625081
Yes 0.163863 0.001551 19.190588 69.808518Now, suppose you wanted to apply potentially different functions to one or more of the columns. To do this, pass a dictionary to agg that contains a mapping of column names to any of the function specifications listed so far:
In [79]: grouped.agg({"tip" : np.max, "size" : "sum"})
Out[79]:
tip size
day smoker
Fri No 3.50 9
Yes 4.73 31
Sat No 9.00 115
Yes 10.00 104
Sun No 6.00 167
Yes 6.50 49
Thur No 6.70 112
Yes 5.00 40
In [80]: grouped.agg({"tip_pct" : ["min", "max", "mean", "std"],
....: "size" : "sum"})
Out[80]:
tip_pct size
min max mean std sum
day smoker
Fri No 0.120385 0.187735 0.151650 0.028123 9
Yes 0.103555 0.263480 0.174783 0.051293 31
Sat No 0.056797 0.291990 0.158048 0.039767 115
Yes 0.035638 0.325733 0.147906 0.061375 104
Sun No 0.059447 0.252672 0.160113 0.042347 167
Yes 0.065660 0.710345 0.187250 0.154134 49
Thur No 0.072961 0.266312 0.160298 0.038774 112
Yes 0.090014 0.241255 0.163863 0.039389 40A DataFrame will have hierarchical columns only if multiple functions are applied to at least one column.
Returning Aggregated Data Without Row Indexes
In all of the examples up until now, the aggregated data comes back with an index, potentially hierarchical, composed from the unique group key combinations. Since this isn’t always desirable, you can disable this behavior in most cases by passing as_index=False to groupby:
In [81]: grouped = tips.groupby(["day", "smoker"], as_index=False)
In [82]: grouped.mean(numeric_only=True)
Out[82]:
day smoker total_bill tip size tip_pct
0 Fri No 18.420000 2.812500 2.250000 0.151650
1 Fri Yes 16.813333 2.714000 2.066667 0.174783
2 Sat No 19.661778 3.102889 2.555556 0.158048
3 Sat Yes 21.276667 2.875476 2.476190 0.147906
4 Sun No 20.506667 3.167895 2.929825 0.160113
5 Sun Yes 24.120000 3.516842 2.578947 0.187250
6 Thur No 17.113111 2.673778 2.488889 0.160298
7 Thur Yes 19.190588 3.030000 2.352941 0.163863Of course, it’s always possible to obtain the result in this format by calling reset_index on the result. Using the as_index=False argument avoids some unnecessary computations.
10.3 Apply: General split-apply-combine
The most general-purpose GroupBy method is apply, which is the subject of this section. apply splits the object being manipulated into pieces, invokes the passed function on each piece, and then attempts to concatenate the pieces.
Returning to the tipping dataset from before, suppose you wanted to select the top five tip_pct values by group. First, write a function that selects the rows with the largest values in a particular column:
In [83]: def top(df, n=5, column="tip_pct"):
....: return df.sort_values(column, ascending=False)[:n]
In [84]: top(tips, n=6)
Out[84]:
total_bill tip smoker day time size tip_pct
172 7.25 5.15 Yes Sun Dinner 2 0.710345
178 9.60 4.00 Yes Sun Dinner 2 0.416667
67 3.07 1.00 Yes Sat Dinner 1 0.325733
232 11.61 3.39 No Sat Dinner 2 0.291990
183 23.17 6.50 Yes Sun Dinner 4 0.280535
109 14.31 4.00 Yes Sat Dinner 2 0.279525Now, if we group by smoker, say, and call apply with this function, we get the following:
In [85]: tips.groupby("smoker").apply(top)
Out[85]:
total_bill tip smoker day time size tip_pct
smoker
No 232 11.61 3.39 No Sat Dinner 2 0.291990
149 7.51 2.00 No Thur Lunch 2 0.266312
51 10.29 2.60 No Sun Dinner 2 0.252672
185 20.69 5.00 No Sun Dinner 5 0.241663
88 24.71 5.85 No Thur Lunch 2 0.236746
Yes 172 7.25 5.15 Yes Sun Dinner 2 0.710345
178 9.60 4.00 Yes Sun Dinner 2 0.416667
67 3.07 1.00 Yes Sat Dinner 1 0.325733
183 23.17 6.50 Yes Sun Dinner 4 0.280535
109 14.31 4.00 Yes Sat Dinner 2 0.279525What has happened here? First, the tips DataFrame is split into groups based on the value of smoker. Then the top function is called on each group, and the results of each function call are glued together using pandas.concat, labeling the pieces with the group names. The result therefore has a hierarchical index with an inner level that contains index values from the original DataFrame.
If you pass a function to apply that takes other arguments or keywords, you can pass these after the function:
In [86]: tips.groupby(["smoker", "day"]).apply(top, n=1, column="total_bill")
Out[86]:
total_bill tip smoker day time size tip_pct
smoker day
No Fri 94 22.75 3.25 No Fri Dinner 2 0.142857
Sat 212 48.33 9.00 No Sat Dinner 4 0.186220
Sun 156 48.17 5.00 No Sun Dinner 6 0.103799
Thur 142 41.19 5.00 No Thur Lunch 5 0.121389
Yes Fri 95 40.17 4.73 Yes Fri Dinner 4 0.117750
Sat 170 50.81 10.00 Yes Sat Dinner 3 0.196812
Sun 182 45.35 3.50 Yes Sun Dinner 3 0.077178
Thur 197 43.11 5.00 Yes Thur Lunch 4 0.115982Beyond these basic usage mechanics, getting the most out of apply may require some creativity. What occurs inside the function passed is up to you; it must either return a pandas object or a scalar value. The rest of this chapter will consist mainly of examples showing you how to solve various problems using groupby.
For example, you may recall that I earlier called describe on a GroupBy object:
In [87]: result = tips.groupby("smoker")["tip_pct"].describe()
In [88]: result
Out[88]:
count mean std min 25% 50% 75%
smoker
No 151.0 0.159328 0.039910 0.056797 0.136906 0.155625 0.185014 \
Yes 93.0 0.163196 0.085119 0.035638 0.106771 0.153846 0.195059
max
smoker
No 0.291990
Yes 0.710345
In [89]: result.unstack("smoker")
Out[89]:
smoker
count No 151.000000
Yes 93.000000
mean No 0.159328
Yes 0.163196
std No 0.039910
Yes 0.085119
min No 0.056797
Yes 0.035638
25% No 0.136906
Yes 0.106771
50% No 0.155625
Yes 0.153846
75% No 0.185014
Yes 0.195059
max No 0.291990
Yes 0.710345
dtype: float64Inside GroupBy, when you invoke a method like describe, it is actually just a shortcut for:
def f(group):
return group.describe()
grouped.apply(f)Suppressing the Group Keys
In the preceding examples, you see that the resulting object has a hierarchical index formed from the group keys, along with the indexes of each piece of the original object. You can disable this by passing group_keys=False to groupby:
In [90]: tips.groupby("smoker", group_keys=False).apply(top)
Out[90]:
total_bill tip smoker day time size tip_pct
232 11.61 3.39 No Sat Dinner 2 0.291990
149 7.51 2.00 No Thur Lunch 2 0.266312
51 10.29 2.60 No Sun Dinner 2 0.252672
185 20.69 5.00 No Sun Dinner 5 0.241663
88 24.71 5.85 No Thur Lunch 2 0.236746
172 7.25 5.15 Yes Sun Dinner 2 0.710345
178 9.60 4.00 Yes Sun Dinner 2 0.416667
67 3.07 1.00 Yes Sat Dinner 1 0.325733
183 23.17 6.50 Yes Sun Dinner 4 0.280535
109 14.31 4.00 Yes Sat Dinner 2 0.279525Quantile and Bucket Analysis
As you may recall from Ch 8: Data Wrangling: Join, Combine, and Reshape, pandas has some tools, in particular pandas.cut and pandas.qcut, for slicing data up into buckets with bins of your choosing, or by sample quantiles. Combining these functions with groupby makes it convenient to perform bucket or quantile analysis on a dataset. Consider a simple random dataset and an equal-length bucket categorization using pandas.cut:
In [91]: frame = pd.DataFrame({"data1": np.random.standard_normal(1000),
....: "data2": np.random.standard_normal(1000)})
In [92]: frame.head()
Out[92]:
data1 data2
0 -0.660524 -0.612905
1 0.862580 0.316447
2 -0.010032 0.838295
3 0.050009 -1.034423
4 0.670216 0.434304
In [93]: quartiles = pd.cut(frame["data1"], 4)
In [94]: quartiles.head(10)
Out[94]:
0 (-1.23, 0.489]
1 (0.489, 2.208]
2 (-1.23, 0.489]
3 (-1.23, 0.489]
4 (0.489, 2.208]
5 (0.489, 2.208]
6 (-1.23, 0.489]
7 (-1.23, 0.489]
8 (-2.956, -1.23]
9 (-1.23, 0.489]
Name: data1, dtype: category
Categories (4, interval[float64, right]): [(-2.956, -1.23] < (-1.23, 0.489] < (0.
489, 2.208] <
(2.208, 3.928]]The Categorical object returned by cut can be passed directly to groupby. So we could compute a set of group statistics for the quartiles, like so:
In [95]: def get_stats(group):
....: return pd.DataFrame(
....: {"min": group.min(), "max": group.max(),
....: "count": group.count(), "mean": group.mean()}
....: )
In [96]: grouped = frame.groupby(quartiles)
In [97]: grouped.apply(get_stats)
Out[97]:
min max count mean
data1
(-2.956, -1.23] data1 -2.949343 -1.230179 94 -1.658818
data2 -3.399312 1.670835 94 -0.033333
(-1.23, 0.489] data1 -1.228918 0.488675 598 -0.329524
data2 -2.989741 3.260383 598 -0.002622
(0.489, 2.208] data1 0.489965 2.200997 298 1.065727
data2 -3.745356 2.954439 298 0.078249
(2.208, 3.928] data1 2.212303 3.927528 10 2.644253
data2 -1.929776 1.765640 10 0.024750Keep in mind the same result could have been computed more simply with:
In [98]: grouped.agg(["min", "max", "count", "mean"])
Out[98]:
data1 data2
min max count mean min max count
data1
(-2.956, -1.23] -2.949343 -1.230179 94 -1.658818 -3.399312 1.670835 94 \
(-1.23, 0.489] -1.228918 0.488675 598 -0.329524 -2.989741 3.260383 598
(0.489, 2.208] 0.489965 2.200997 298 1.065727 -3.745356 2.954439 298
(2.208, 3.928] 2.212303 3.927528 10 2.644253 -1.929776 1.765640 10
mean
data1
(-2.956, -1.23] -0.033333
(-1.23, 0.489] -0.002622
(0.489, 2.208] 0.078249
(2.208, 3.928] 0.024750 These were equal-length buckets; to compute equal-size buckets based on sample quantiles, use pandas.qcut. We can pass 4 as the number of bucket compute sample quartiles, and pass labels=False to obtain just the quartile indices instead of intervals:
In [99]: quartiles_samp = pd.qcut(frame["data1"], 4, labels=False)
In [100]: quartiles_samp.head()
Out[100]:
0 1
1 3
2 2
3 2
4 3
Name: data1, dtype: int64
In [101]: grouped = frame.groupby(quartiles_samp)
In [102]: grouped.apply(get_stats)
Out[102]:
min max count mean
data1
0 data1 -2.949343 -0.685484 250 -1.212173
data2 -3.399312 2.628441 250 -0.027045
1 data1 -0.683066 -0.030280 250 -0.368334
data2 -2.630247 3.260383 250 -0.027845
2 data1 -0.027734 0.618965 250 0.295812
data2 -3.056990 2.458842 250 0.014450
3 data1 0.623587 3.927528 250 1.248875
data2 -3.745356 2.954439 250 0.115899Example: Filling Missing Values with Group-Specific Values
When cleaning up missing data, in some cases you will remove data observations using dropna, but in others you may want to fill in the null (NA) values using a fixed value or some value derived from the data. fillna is the right tool to use; for example, here I fill in the null values with the mean:
In [103]: s = pd.Series(np.random.standard_normal(6))
In [104]: s[::2] = np.nan
In [105]: s
Out[105]:
0 NaN
1 0.227290
2 NaN
3 -2.153545
4 NaN
5 -0.375842
dtype: float64
In [106]: s.fillna(s.mean())
Out[106]:
0 -0.767366
1 0.227290
2 -0.767366
3 -2.153545
4 -0.767366
5 -0.375842
dtype: float64Suppose you need the fill value to vary by group. One way to do this is to group the data and use apply with a function that calls fillna on each data chunk. Here is some sample data on US states divided into eastern and western regions:
In [107]: states = ["Ohio", "New York", "Vermont", "Florida",
.....: "Oregon", "Nevada", "California", "Idaho"]
In [108]: group_key = ["East", "East", "East", "East",
.....: "West", "West", "West", "West"]
In [109]: data = pd.Series(np.random.standard_normal(8), index=states)
In [110]: data
Out[110]:
Ohio 0.329939
New York 0.981994
Vermont 1.105913
Florida -1.613716
Oregon 1.561587
Nevada 0.406510
California 0.359244
Idaho -0.614436
dtype: float64Let's set some values in the data to be missing:
In [111]: data[["Vermont", "Nevada", "Idaho"]] = np.nan
In [112]: data
Out[112]:
Ohio 0.329939
New York 0.981994
Vermont NaN
Florida -1.613716
Oregon 1.561587
Nevada NaN
California 0.359244
Idaho NaN
dtype: float64
In [113]: data.groupby(group_key).size()
Out[113]:
East 4
West 4
dtype: int64
In [114]: data.groupby(group_key).count()
Out[114]:
East 3
West 2
dtype: int64
In [115]: data.groupby(group_key).mean()
Out[115]:
East -0.100594
West 0.960416
dtype: float64We can fill the NA values using the group means, like so:
In [116]: def fill_mean(group):
.....: return group.fillna(group.mean())
In [117]: data.groupby(group_key).apply(fill_mean)
Out[117]:
East Ohio 0.329939
New York 0.981994
Vermont -0.100594
Florida -1.613716
West Oregon 1.561587
Nevada 0.960416
California 0.359244
Idaho 0.960416
dtype: float64In another case, you might have predefined fill values in your code that vary by group. Since the groups have a name attribute set internally, we can use that:
In [118]: fill_values = {"East": 0.5, "West": -1}
In [119]: def fill_func(group):
.....: return group.fillna(fill_values[group.name])
In [120]: data.groupby(group_key).apply(fill_func)
Out[120]:
East Ohio 0.329939
New York 0.981994
Vermont 0.500000
Florida -1.613716
West Oregon 1.561587
Nevada -1.000000
California 0.359244
Idaho -1.000000
dtype: float64Example: Random Sampling and Permutation
Suppose you wanted to draw a random sample (with or without replacement) from a large dataset for Monte Carlo simulation purposes or some other application. There are a number of ways to perform the “draws”; here we use the sample method for Series.
To demonstrate, here’s a way to construct a deck of English-style playing cards:
suits = ["H", "S", "C", "D"] # Hearts, Spades, Clubs, Diamonds
card_val = (list(range(1, 11)) + [10] * 3) * 4
base_names = ["A"] + list(range(2, 11)) + ["J", "K", "Q"]
cards = []
for suit in suits:
cards.extend(str(num) + suit for num in base_names)
deck = pd.Series(card_val, index=cards)Now we have a Series of length 52 whose index contains card names, and values are the ones used in blackjack and other games (to keep things simple, I let the ace "A" be 1):
In [122]: deck.head(13)
Out[122]:
AH 1
2H 2
3H 3
4H 4
5H 5
6H 6
7H 7
8H 8
9H 9
10H 10
JH 10
KH 10
QH 10
dtype: int64Now, based on what I said before, drawing a hand of five cards from the deck could be written as:
In [123]: def draw(deck, n=5):
.....: return deck.sample(n)
In [124]: draw(deck)
Out[124]:
4D 4
QH 10
8S 8
7D 7
9C 9
dtype: int64Suppose you wanted two random cards from each suit. Because the suit is the last character of each card name, we can group based on this and use apply:
In [125]: def get_suit(card):
.....: # last letter is suit
.....: return card[-1]
In [126]: deck.groupby(get_suit).apply(draw, n=2)
Out[126]:
C 6C 6
KC 10
D 7D 7
3D 3
H 7H 7
9H 9
S 2S 2
QS 10
dtype: int64Alternatively, we could pass group_keys=False to drop the outer suit index, leaving in just the selected cards:
In [127]: deck.groupby(get_suit, group_keys=False).apply(draw, n=2)
Out[127]:
AC 1
3C 3
5D 5
4D 4
10H 10
7H 7
QS 10
7S 7
dtype: int64Example: Group Weighted Average and Correlation
Under the split-apply-combine paradigm of groupby, operations between columns in a DataFrame or two Series, such as a group weighted average, are possible. As an example, take this dataset containing group keys, values, and some weights:
In [128]: df = pd.DataFrame({"category": ["a", "a", "a", "a",
.....: "b", "b", "b", "b"],
.....: "data": np.random.standard_normal(8),
.....: "weights": np.random.uniform(size=8)})
In [129]: df
Out[129]:
category data weights
0 a -1.691656 0.955905
1 a 0.511622 0.012745
2 a -0.401675 0.137009
3 a 0.968578 0.763037
4 b -1.818215 0.492472
5 b 0.279963 0.832908
6 b -0.200819 0.658331
7 b -0.217221 0.612009The weighted average by category would then be:
In [130]: grouped = df.groupby("category")
In [131]: def get_wavg(group):
.....: return np.average(group["data"], weights=group["weights"])
In [132]: grouped.apply(get_wavg)
Out[132]:
category
a -0.495807
b -0.357273
dtype: float64As another example, consider a financial dataset originally obtained from Yahoo! Finance containing end-of-day prices for a few stocks and the S&P 500 index (the SPX symbol):
In [133]: close_px = pd.read_csv("examples/stock_px.csv", parse_dates=True,
.....: index_col=0)
In [134]: close_px.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 2214 entries, 2003-01-02 to 2011-10-14
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AAPL 2214 non-null float64
1 MSFT 2214 non-null float64
2 XOM 2214 non-null float64
3 SPX 2214 non-null float64
dtypes: float64(4)
memory usage: 86.5 KB
In [135]: close_px.tail(4)
Out[135]:
AAPL MSFT XOM SPX
2011-10-11 400.29 27.00 76.27 1195.54
2011-10-12 402.19 26.96 77.16 1207.25
2011-10-13 408.43 27.18 76.37 1203.66
2011-10-14 422.00 27.27 78.11 1224.58The DataFrame info() method here is a convenient way to get an overview of the contents of a DataFrame.
One task of interest might be to compute a DataFrame consisting of the yearly correlations of daily returns (computed from percent changes) with SPX. As one way to do this, we first create a function that computes the pair-wise correlation of each column with the "SPX" column:
In [136]: def spx_corr(group):
.....: return group.corrwith(group["SPX"])Next, we compute percent change on close_px using pct_change:
In [137]: rets = close_px.pct_change().dropna()Lastly, we group these percent changes by year, which can be extracted from each row label with a one-line function that returns the year attribute of each datetime label:
In [138]: def get_year(x):
.....: return x.year
In [139]: by_year = rets.groupby(get_year)
In [140]: by_year.apply(spx_corr)
Out[140]:
AAPL MSFT XOM SPX
2003 0.541124 0.745174 0.661265 1.0
2004 0.374283 0.588531 0.557742 1.0
2005 0.467540 0.562374 0.631010 1.0
2006 0.428267 0.406126 0.518514 1.0
2007 0.508118 0.658770 0.786264 1.0
2008 0.681434 0.804626 0.828303 1.0
2009 0.707103 0.654902 0.797921 1.0
2010 0.710105 0.730118 0.839057 1.0
2011 0.691931 0.800996 0.859975 1.0You could also compute intercolumn correlations. Here we compute the annual correlation between Apple and Microsoft:
In [141]: def corr_aapl_msft(group):
.....: return group["AAPL"].corr(group["MSFT"])
In [142]: by_year.apply(corr_aapl_msft)
Out[142]:
2003 0.480868
2004 0.259024
2005 0.300093
2006 0.161735
2007 0.417738
2008 0.611901
2009 0.432738
2010 0.571946
2011 0.581987
dtype: float64Example: Group-Wise Linear Regression
In the same theme as the previous example, you can use groupby to perform more complex group-wise statistical analysis, as long as the function returns a pandas object or scalar value. For example, I can define the following regress function (using the statsmodels econometrics library), which executes an ordinary least squares (OLS) regression on each chunk of data:
import statsmodels.api as sm
def regress(data, yvar=None, xvars=None):
Y = data[yvar]
X = data[xvars]
X["intercept"] = 1.
result = sm.OLS(Y, X).fit()
return result.paramsYou can install statsmodels with conda if you don't have it already:
conda install statsmodelsNow, to run a yearly linear regression of AAPL on SPX returns, execute:
In [144]: by_year.apply(regress, yvar="AAPL", xvars=["SPX"])
Out[144]:
SPX intercept
2003 1.195406 0.000710
2004 1.363463 0.004201
2005 1.766415 0.003246
2006 1.645496 0.000080
2007 1.198761 0.003438
2008 0.968016 -0.001110
2009 0.879103 0.002954
2010 1.052608 0.001261
2011 0.806605 0.00151410.4 Group Transforms and "Unwrapped" GroupBys
In Apply: General split-apply-combine, we looked at the apply method in grouped operations for performing transformations. There is another built-in method called transform, which is similar to apply but imposes more constraints on the kind of function you can use:
It can produce a scalar value to be broadcast to the shape of the group.
It can produce an object of the same shape as the input group.
It must not mutate its input.
Let's consider a simple example for illustration:
In [145]: df = pd.DataFrame({'key': ['a', 'b', 'c'] * 4,
.....: 'value': np.arange(12.)})
In [146]: df
Out[146]:
key value
0 a 0.0
1 b 1.0
2 c 2.0
3 a 3.0
4 b 4.0
5 c 5.0
6 a 6.0
7 b 7.0
8 c 8.0
9 a 9.0
10 b 10.0
11 c 11.0Here are the group means by key:
In [147]: g = df.groupby('key')['value']
In [148]: g.mean()
Out[148]:
key
a 4.5
b 5.5
c 6.5
Name: value, dtype: float64Suppose instead we wanted to produce a Series of the same shape as df['value'] but with values replaced by the average grouped by 'key'. We can pass a function that computes the mean of a single group to transform:
In [149]: def get_mean(group):
.....: return group.mean()
In [150]: g.transform(get_mean)
Out[150]:
0 4.5
1 5.5
2 6.5
3 4.5
4 5.5
5 6.5
6 4.5
7 5.5
8 6.5
9 4.5
10 5.5
11 6.5
Name: value, dtype: float64For built-in aggregation functions, we can pass a string alias as with the GroupBy agg method:
In [151]: g.transform('mean')
Out[151]:
0 4.5
1 5.5
2 6.5
3 4.5
4 5.5
5 6.5
6 4.5
7 5.5
8 6.5
9 4.5
10 5.5
11 6.5
Name: value, dtype: float64Like apply, transform works with functions that return Series, but the result must be the same size as the input. For example, we can multiply each group by 2 using a helper function:
In [152]: def times_two(group):
.....: return group * 2
In [153]: g.transform(times_two)
Out[153]:
0 0.0
1 2.0
2 4.0
3 6.0
4 8.0
5 10.0
6 12.0
7 14.0
8 16.0
9 18.0
10 20.0
11 22.0
Name: value, dtype: float64As a more complicated example, we can compute the ranks in descending order for each group:
In [154]: def get_ranks(group):
.....: return group.rank(ascending=False)
In [155]: g.transform(get_ranks)
Out[155]:
0 4.0
1 4.0
2 4.0
3 3.0
4 3.0
5 3.0
6 2.0
7 2.0
8 2.0
9 1.0
10 1.0
11 1.0
Name: value, dtype: float64Consider a group transformation function composed from simple aggregations:
In [156]: def normalize(x):
.....: return (x - x.mean()) / x.std()We can obtain equivalent results in this case using either transform or apply:
In [157]: g.transform(normalize)
Out[157]:
0 -1.161895
1 -1.161895
2 -1.161895
3 -0.387298
4 -0.387298
5 -0.387298
6 0.387298
7 0.387298
8 0.387298
9 1.161895
10 1.161895
11 1.161895
Name: value, dtype: float64
In [158]: g.apply(normalize)
Out[158]:
key
a 0 -1.161895
3 -0.387298
6 0.387298
9 1.161895
b 1 -1.161895
4 -0.387298
7 0.387298
10 1.161895
c 2 -1.161895
5 -0.387298
8 0.387298
11 1.161895
Name: value, dtype: float64Built-in aggregate functions like 'mean' or 'sum' are often much faster than a general apply function. These also have a "fast path" when used with transform. This allows us to perform what is called an unwrapped group operation:
In [159]: g.transform('mean')
Out[159]:
0 4.5
1 5.5
2 6.5
3 4.5
4 5.5
5 6.5
6 4.5
7 5.5
8 6.5
9 4.5
10 5.5
11 6.5
Name: value, dtype: float64
In [160]: normalized = (df['value'] - g.transform('mean')) / g.transform('std')
In [161]: normalized
Out[161]:
0 -1.161895
1 -1.161895
2 -1.161895
3 -0.387298
4 -0.387298
5 -0.387298
6 0.387298
7 0.387298
8 0.387298
9 1.161895
10 1.161895
11 1.161895
Name: value, dtype: float64Here, we are doing arithmetic between the outputs of multiple GroupBy operations instead of writing a function and passing it to groupby(...).apply. That is what is meant by "unwrapped."
While an unwrapped group operation may involve multiple group aggregations, the overall benefit of vectorized operations often outweighs this.
10.5 Pivot Tables and Cross-Tabulation
A pivot table is a data summarization tool frequently found in spreadsheet programs and other data analysis software. It aggregates a table of data by one or more keys, arranging the data in a rectangle with some of the group keys along the rows and some along the columns. Pivot tables in Python with pandas are made possible through the groupby facility described in this chapter, combined with reshape operations utilizing hierarchical indexing. DataFrame also has a pivot_table method, and there is also a top-level pandas.pivot_table function. In addition to providing a convenience interface to groupby, pivot_table can add partial totals, also known as margins.
Returning to the tipping dataset, suppose you wanted to compute a table of group means (the default pivot_table aggregation type) arranged by day and smoker on the rows:
In [162]: tips.head()
Out[162]:
total_bill tip smoker day time size tip_pct
0 16.99 1.01 No Sun Dinner 2 0.059447
1 10.34 1.66 No Sun Dinner 3 0.160542
2 21.01 3.50 No Sun Dinner 3 0.166587
3 23.68 3.31 No Sun Dinner 2 0.139780
4 24.59 3.61 No Sun Dinner 4 0.146808
In [163]: tips.pivot_table(index=["day", "smoker"],
.....: values=["size", "tip", "tip_pct", "total_bill"])
Out[163]:
size tip tip_pct total_bill
day smoker
Fri No 2.250000 2.812500 0.151650 18.420000
Yes 2.066667 2.714000 0.174783 16.813333
Sat No 2.555556 3.102889 0.158048 19.661778
Yes 2.476190 2.875476 0.147906 21.276667
Sun No 2.929825 3.167895 0.160113 20.506667
Yes 2.578947 3.516842 0.187250 24.120000
Thur No 2.488889 2.673778 0.160298 17.113111
Yes 2.352941 3.030000 0.163863 19.190588This could have been produced with groupby directly, using tips.groupby(["day", "smoker"]).mean(). Now, suppose we want to take the average of only tip_pct and size, and additionally group by time. I’ll put smoker in the table columns and time and day in the rows:
In [164]: tips.pivot_table(index=["time", "day"], columns="smoker",
.....: values=["tip_pct", "size"])
Out[164]:
size tip_pct
smoker No Yes No Yes
time day
Dinner Fri 2.000000 2.222222 0.139622 0.165347
Sat 2.555556 2.476190 0.158048 0.147906
Sun 2.929825 2.578947 0.160113 0.187250
Thur 2.000000 NaN 0.159744 NaN
Lunch Fri 3.000000 1.833333 0.187735 0.188937
Thur 2.500000 2.352941 0.160311 0.163863We could augment this table to include partial totals by passing margins=True. This has the effect of adding All row and column labels, with corresponding values being the group statistics for all the data within a single tier:
In [165]: tips.pivot_table(index=["time", "day"], columns="smoker",
.....: values=["tip_pct", "size"], margins=True)
Out[165]:
size tip_pct
smoker No Yes All No Yes All
time day
Dinner Fri 2.000000 2.222222 2.166667 0.139622 0.165347 0.158916
Sat 2.555556 2.476190 2.517241 0.158048 0.147906 0.153152
Sun 2.929825 2.578947 2.842105 0.160113 0.187250 0.166897
Thur 2.000000 NaN 2.000000 0.159744 NaN 0.159744
Lunch Fri 3.000000 1.833333 2.000000 0.187735 0.188937 0.188765
Thur 2.500000 2.352941 2.459016 0.160311 0.163863 0.161301
All 2.668874 2.408602 2.569672 0.159328 0.163196 0.160803Here, the All values are means without taking into account smoker versus non-smoker (the All columns) or any of the two levels of grouping on the rows (the All row).
To use an aggregation function other than mean, pass it to the aggfunc keyword argument. For example, "count" or len will give you a cross-tabulation (count or frequency) of group sizes (though "count" will exclude null values from the count within data groups, while len will not):
In [166]: tips.pivot_table(index=["time", "smoker"], columns="day",
.....: values="tip_pct", aggfunc=len, margins=True)
Out[166]:
day Fri Sat Sun Thur All
time smoker
Dinner No 3.0 45.0 57.0 1.0 106
Yes 9.0 42.0 19.0 NaN 70
Lunch No 1.0 NaN NaN 44.0 45
Yes 6.0 NaN NaN 17.0 23
All 19.0 87.0 76.0 62.0 244If some combinations are empty (or otherwise NA), you may wish to pass a fill_value:
In [167]: tips.pivot_table(index=["time", "size", "smoker"], columns="day",
.....: values="tip_pct", fill_value=0)
Out[167]:
day Fri Sat Sun Thur
time size smoker
Dinner 1 No 0.000000 0.137931 0.000000 0.000000
Yes 0.000000 0.325733 0.000000 0.000000
2 No 0.139622 0.162705 0.168859 0.159744
Yes 0.171297 0.148668 0.207893 0.000000
3 No 0.000000 0.154661 0.152663 0.000000
... ... ... ... ...
Lunch 3 Yes 0.000000 0.000000 0.000000 0.204952
4 No 0.000000 0.000000 0.000000 0.138919
Yes 0.000000 0.000000 0.000000 0.155410
5 No 0.000000 0.000000 0.000000 0.121389
6 No 0.000000 0.000000 0.000000 0.173706
[21 rows x 4 columns]See Table 10.2 for a summary of pivot_table options.
| Argument | Description |
|---|---|
values |
Column name or names to aggregate; by default, aggregates all numeric columns |
index |
Column names or other group keys to group on the rows of the resulting pivot table |
columns |
Column names or other group keys to group on the columns of the resulting pivot table |
aggfunc |
Aggregation function or list of functions ("mean" by default); can be any function valid in a groupby context |
fill_value |
Replace missing values in the result table |
dropna |
If True, do not include columns whose entries are all NA |
margins |
Add row/column subtotals and grand total (False by default) |
margins_name |
Name to use for the margin row/column labels when passing margins=True; defaults to "All" |
observed |
With Categorical group keys, if True, show only the observed category values in the keys rather than all categories |
Cross-Tabulations: Crosstab
A cross-tabulation (or crosstab for short) is a special case of a pivot table that computes group frequencies. Here is an example:
In [168]: from io import StringIO
In [169]: data = """Sample Nationality Handedness
.....: 1 USA Right-handed
.....: 2 Japan Left-handed
.....: 3 USA Right-handed
.....: 4 Japan Right-handed
.....: 5 Japan Left-handed
.....: 6 Japan Right-handed
.....: 7 USA Right-handed
.....: 8 USA Left-handed
.....: 9 Japan Right-handed
.....: 10 USA Right-handed"""
.....:
In [170]: data = pd.read_table(StringIO(data), sep="\s+")In [171]: data
Out[171]:
Sample Nationality Handedness
0 1 USA Right-handed
1 2 Japan Left-handed
2 3 USA Right-handed
3 4 Japan Right-handed
4 5 Japan Left-handed
5 6 Japan Right-handed
6 7 USA Right-handed
7 8 USA Left-handed
8 9 Japan Right-handed
9 10 USA Right-handedAs part of some survey analysis, we might want to summarize this data by nationality and handedness. You could use pivot_table to do this, but the pandas.crosstab function can be more convenient:
In [172]: pd.crosstab(data["Nationality"], data["Handedness"], margins=True)
Out[172]:
Handedness Left-handed Right-handed All
Nationality
Japan 2 3 5
USA 1 4 5
All 3 7 10The first two arguments to crosstab can each be an array or Series or a list of arrays. As in the tips data:
In [173]: pd.crosstab([tips["time"], tips["day"]], tips["smoker"], margins=True)
Out[173]:
smoker No Yes All
time day
Dinner Fri 3 9 12
Sat 45 42 87
Sun 57 19 76
Thur 1 0 1
Lunch Fri 1 6 7
Thur 44 17 61
All 151 93 24410.6 Conclusion
Mastering pandas's data grouping tools can help with data cleaning and modeling or statistical analysis work. In Ch 13: Data Analysis Examples we will look at several more example use cases for groupby on real data.
In the next chapter, we turn our attention to time series data.